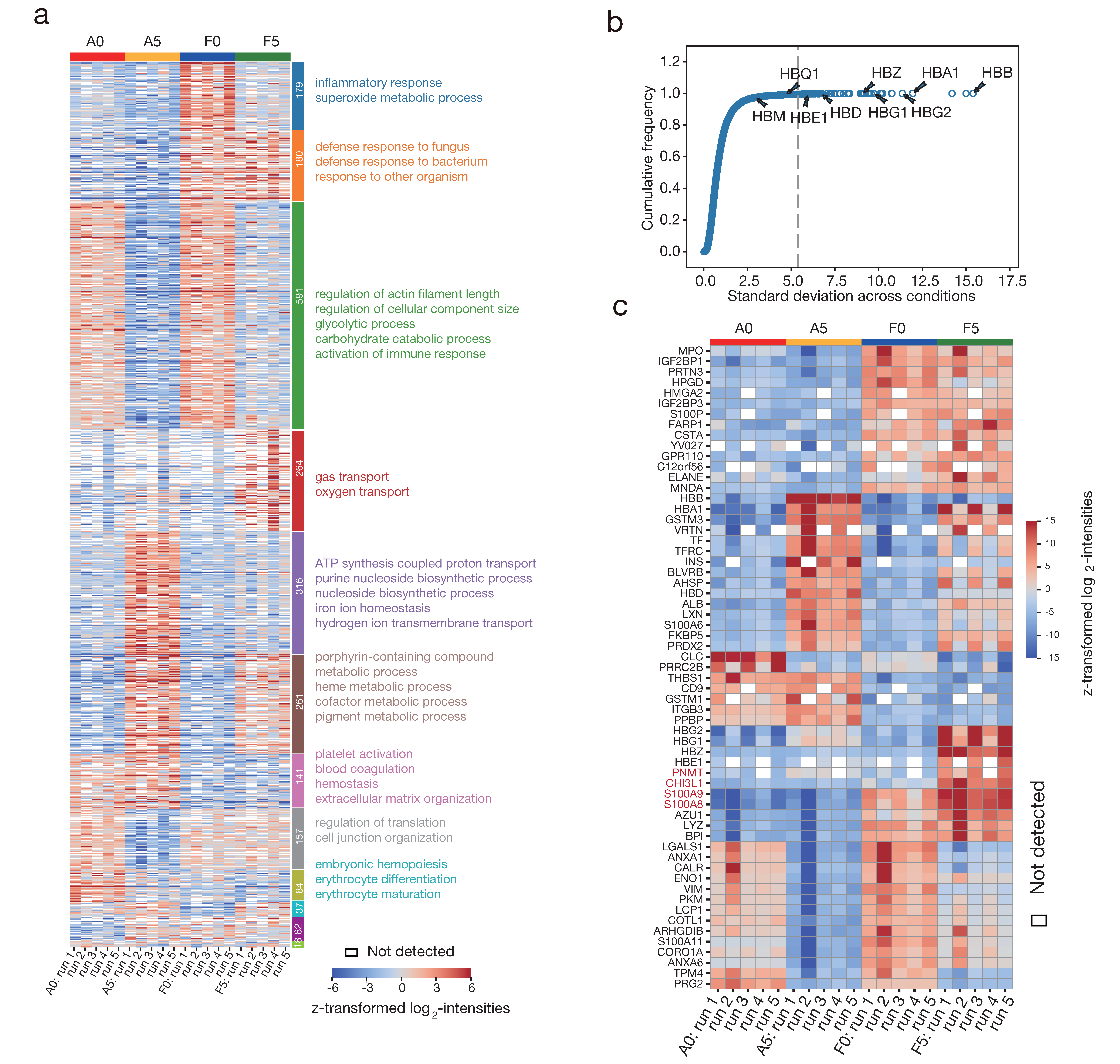
HVPs identification and clustering
The chi-square statistics derived by zMAP along with the associated numbers of degrees of freedom were summed across MS runs for each protein, giving rise to a p-value that assessed the overall expression variability of the protein. This functionality is designed to select hypervariable proteins (HVPs) from the output files of zMAP and cluster them to obtain multiple expression signatures. Then, pathway enrichment analysis was conducted on these signatures to reveal biological insights.