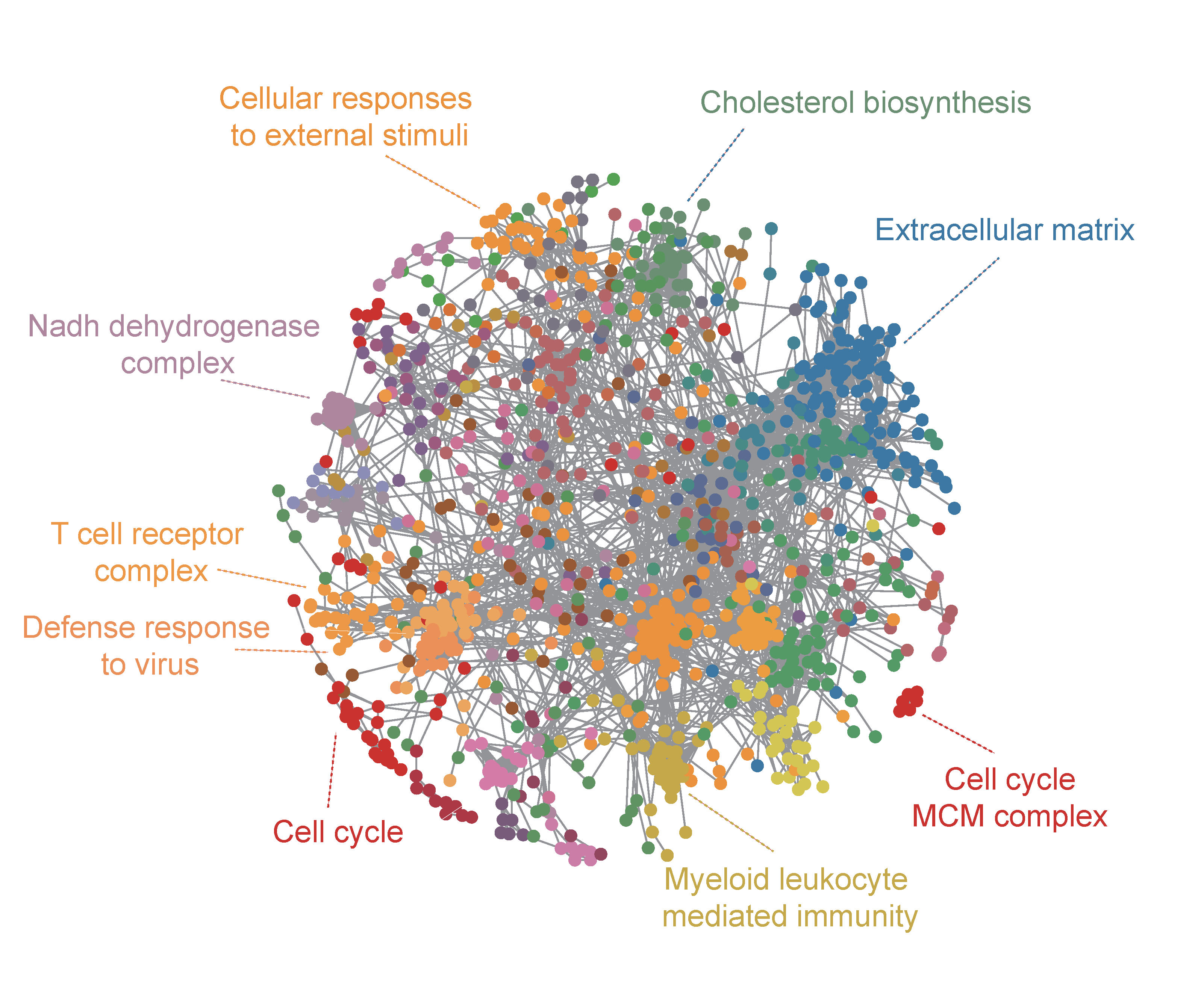
Network analysis
A protein co-expression network is constructed based on the z-statistics. Proteins detected in more than half of the samples are included in the analysis. We first used the KNN method to impute missing values in the z-statistic matrix. Then, we obtained the PTCCs for all pairs of the proteins by applying a previously developed method for covariance matrix estimation. Finally, the statistical significance of each PTCC was assessed based on a mixture model. When constructing the co-expression network, an edge was added to link a protein pair if and only if the corresponding PTCC was positive and significant.