Chinese Spring chromatin states
About chromation state:
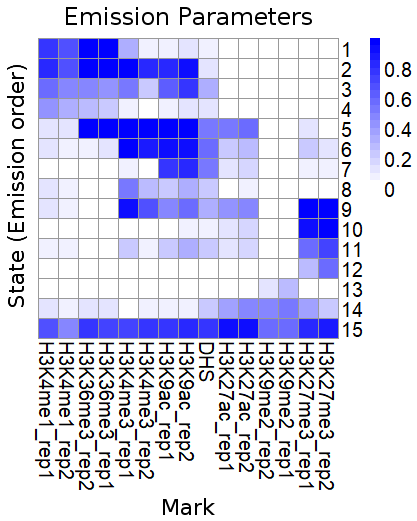
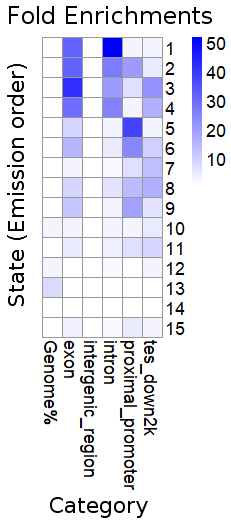
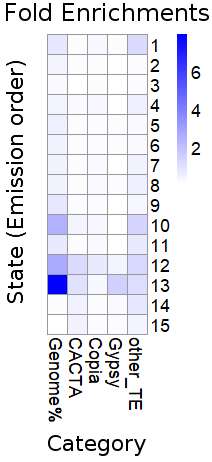
We collected 15 Wheat epigenomic datasets, including ChIP-seq, DNase-seq. A Hidden Markov Model was applied to aggregate or collapse the multi-dimensional matrices into a small number of chromatin states. We identified 15 chromatin states located in 1,714,604 segments across the whole genome and different chromatin states contained different epigenetic features.
After chromatin state partition, we employed self-organization mapping (SOM) to integrate, visualize, and mine diverse epigenomic datasets, including 1350 units that contained different chromatin signatures.
All of 15 states:
| State | Preferential epigenetics marks | Preferential location (Genome) | Preferential location (TE) |
|---|
| 1 | H3K4me1,H3K36me3,H3K4me3 | Exon,Intron | Copia |
| 2 | H3K4me1,H3K36me3,H3K4me3,H3K9ac,acessible DNA | Exon,Intron,Proximal promoter | CACTA,Copia |
| 3 | H3K4me1,H3K36me3,H3K4me3,H3K9ac,acessible DNA | Exon,Intron,TES_Down2k | CACTA,Copia |
| 4 | H3K4me1,H3K36me3 | Exon,Intron,TES_Down2k | CACTA,Copia |
| 5 | H3K36me3,H3K4me3,H3K9ac,acessible DNA,H3K27ac | Proximal promoter | CACTA,Copia,Gypsy |
| 6 | H3K4me3,H3K9ac,acessible DNA | Exon,Proximal promoter,TES_Down2k | CACTA,Copia,Gypsy |
| 7 | H3K9ac,acessible DNA | Proximal promoter | Copia |
| 8 | H3K4me3,H3K9ac,acessible DNA | Exon,Proximal promoter | Copia |
| 9 | H3K4me3,H3K9ac,acessible DNA,H3K27ac,H3K27me3 | Exon,Proximal promoter | CACTA,Copia |
| 10 | H3K27me3 | Exon,Intron,Proximal promoter | CACTA |
| 11 | H3K9ac,H3K27me3,acessible DNA | Exon,Intron,Proximal promoter | CACTA,Copia |
| 12 | H3K27me3 | Intron | CACTA,Copia |
| 13 | H3K9me2 | Intergenic region | CACTA,Gypsy |
| 14 | H3K27ac,H3K9me2,H3K27me3 | Intergenic Region,Intron,Proximal promoter,TES_Down2k | CACTA |
| 15 | H3K4me1,H3K36me3,H3K4me3,H3K9ac,acessible DNA,H3K27ac,H3K9me2,H3K27me3 | Exon,Proximal promoter,TES_Down2k | CACTA |
Segment counts of SOM map | Nucleotide counts of SOM map |
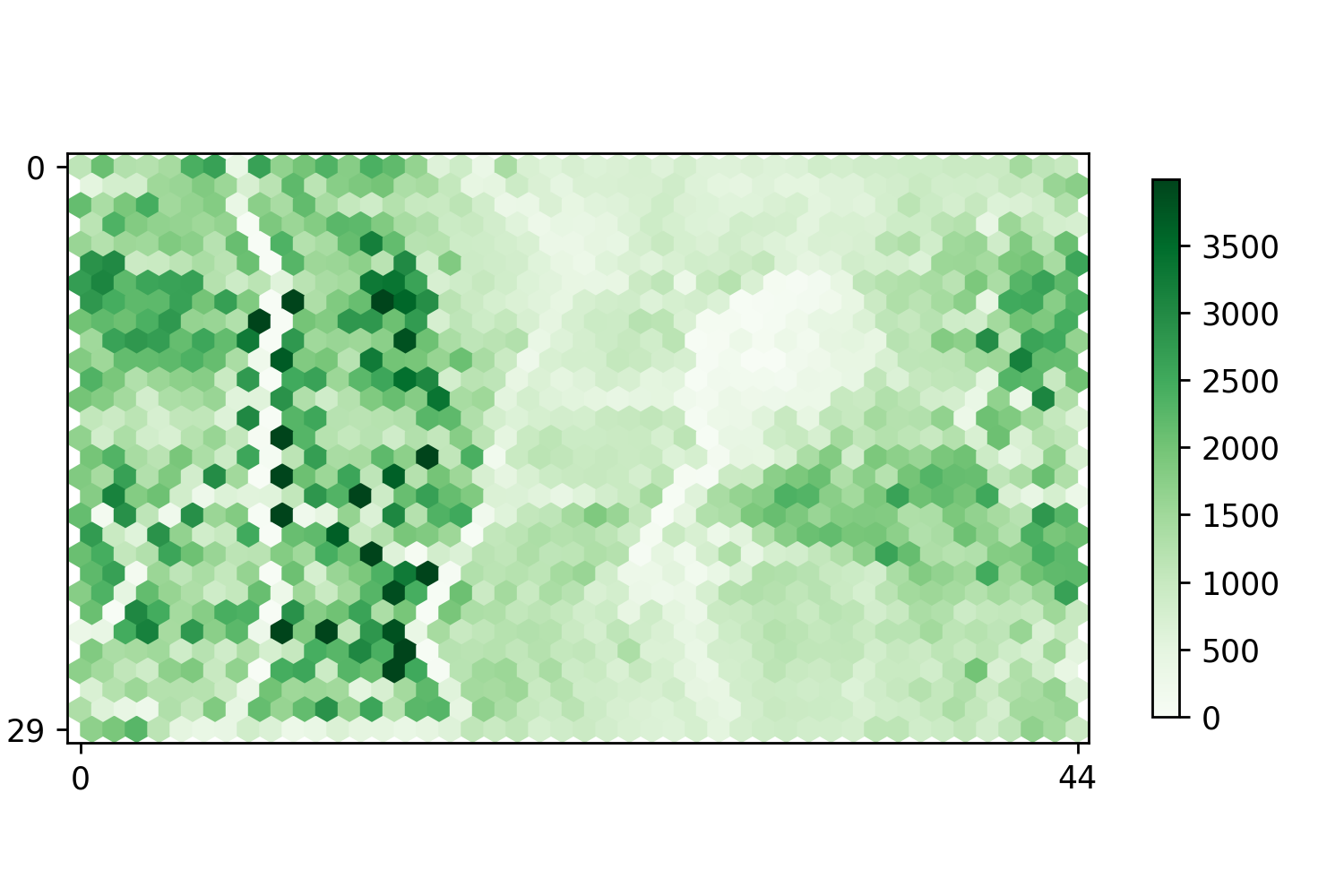 | 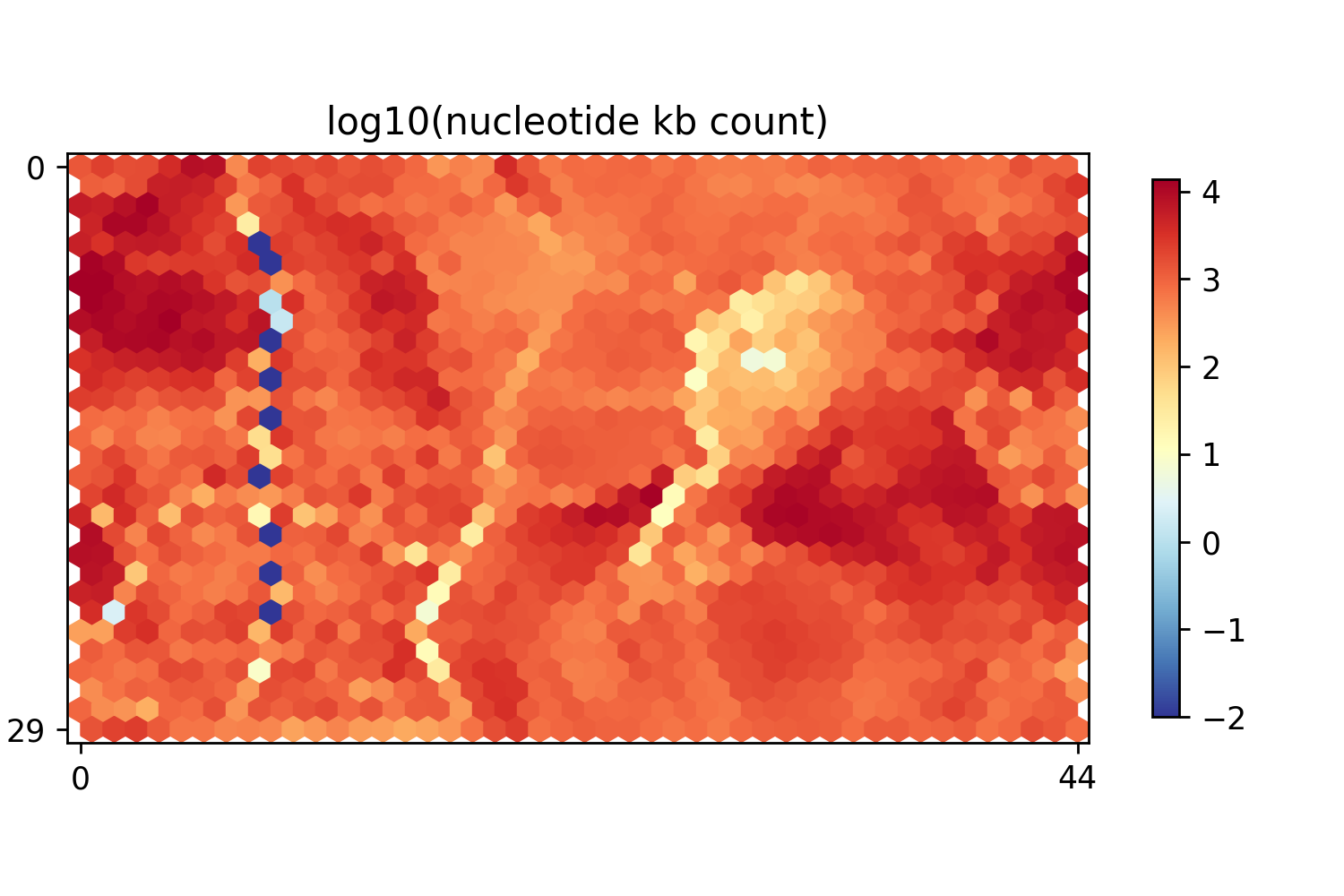 |